I have a soft spot for the Danes. Spending five summers hammering nails with a couple of Danish homebuilders in Ontario taught me the value of being well-read and beer at morning coffee, lunch, and afternoon coffee. My friend John Kierkegaard would say, the beer is nice, but the work, it isn’t really so good.
 When I went to Copenhagen for a scientific meeting, sure enough, there was beer at morning coffee.
When I went to Copenhagen for a scientific meeting, sure enough, there was beer at morning coffee.
The Technical University of Denmark reports that almost every other registered salmonella infection in Denmark in 2014 was brought back by Danes travelling overseas. Travel thus remains the largest cause of salmonella infections. An outbreak of salmonella from Danish eggs was also recorded in 2014, which is the first time in five years and illness was again attributed Danish chicken meat.
These are some of the findings presented in the annual report on the occurrence of diseases that can be transmitted from animals and food to humans. The report was prepared by the National Food Institute, Technical University of Denmark, in cooperation with Statens Serum Institut, the national institute of public health, and the Danish Veterinary and Food Administration.
In 2014 a total of 1,122 salmonella infections were reported among Danes, which is equivalent to 19.9 infected cases per 100,000 inhabitants. The figure is in line with the previous year when a historically low number of Danes was infected with salmonella.
In all, 48% became ill with salmonella after travelling overseas in 2014. Most of those who returned home with a travel-related infection had been to Thailand (17.5%), Turkey (15.4%) and Spain (6.4%).
Thus, foreign travel is still the largest cause of salmonella infections among Danes.
 In the annual source account which the National Food Institute calculates, salmonella infections were attributed Danish chicken meat for the first time since 2011. In total 2% of the infections were estimated to be attributed this source.
In the annual source account which the National Food Institute calculates, salmonella infections were attributed Danish chicken meat for the first time since 2011. In total 2% of the infections were estimated to be attributed this source.
“For two decades Danish producers, authorities and researchers have successfully worked hard to make fresh chicken salmonella-free. It is not allowed to sell fresh meat from Danish chickens if the flock is positive for salmonella. There will always be a small risk that positive fresh meat goes under the surveillance radar and makes its way to store refrigerators. This is why it is important to continue to have a close monitoring,” Senior Academic Officer Birgitte Helwigh from the National Food Institute says.
The first salmonella outbreak from Danish eggs for five years has also been registered in 2014. “It has been five years since we last had a foodborne outbreak caused by Danish eggs. The outbreak was associated with an outbreak of acute salmonella illness in the flock, which is extremely rare. The results show how important it is that producers and authorities continue to focus on maintaining the low incidence of salmonella in the egg production,” says Birgitte Helwigh says.
Danish pork was the food source associated with the most infections among persons infected in Denmark. Overall 15% of the reported illness cases were attributed to Danish pork. There were three outbreaks where Danish pork was registered as the source of infection, which contributed 4.6% of the cases.
Approximately one fifth of all salmonella cases in Denmark were not attributed to a specific food source. The reason may be that the cases were caused by foods which were not included in the salmonella source account, e.g. fruit and vegetables, or other sources of infection such as contact with livestock and pets.
With 3,782 cases registered in 2014, campylobacter still causes the most cases of foodborne bacterial illnesses in Denmark. In 2014 a total of 92 listeria infections were registered, which is an increase of 84% compared to the year before. The increase is mainly due to an outbreak in “rullepølse” (a Danish cold cut ready-to-eat speciality) with 41 reported cases.
In 2014, a total of 60 foodborne disease outbreaks were registered compared with 74 outbreaks the year before. An outbreak is when several people become sick from the same food source. As in previous years, norovirus caused the most outbreaks (40%). These outbreaks usually take place in restaurants, where a total of 363 people were infected in 24 of the recorded outbreaks.
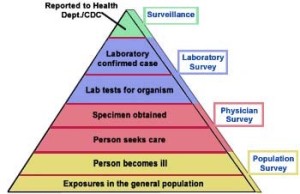
 To better understand the epidemiology of foodborne disease and to direct prevention efforts, we examined incidence of Salmonella infection, Shiga toxin–producing Escherichia coli infection, and hemolytic uremic syndrome by census tract–level socioeconomic status (SES) in the Connecticut Foodborne Diseases Active Surveillance Network site for 2000–2011.
To better understand the epidemiology of foodborne disease and to direct prevention efforts, we examined incidence of Salmonella infection, Shiga toxin–producing Escherichia coli infection, and hemolytic uremic syndrome by census tract–level socioeconomic status (SES) in the Connecticut Foodborne Diseases Active Surveillance Network site for 2000–2011.