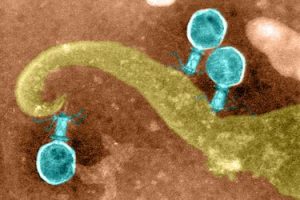
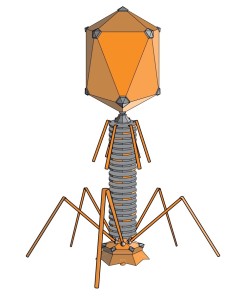
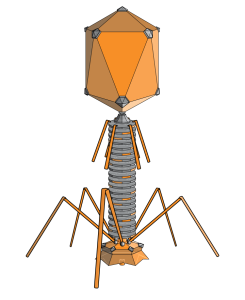
Given that phages are able to destroy bacteria, they are of particular interest to science. Basic researchers from the Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) in Berlin are especially interested in the tube used by phages to implant their DNA (or RNA – dp) into bacteria. In collaboration with colleagues from Forschungszentrum Jülich and Jena University Hospital, they have now revealed the 3D structure of this crucial phage component in atomic resolution. The key to success was combining two methods – solid-state NMR and cryo-electron microscopy. The study has just been published in the journal Nature Communications.
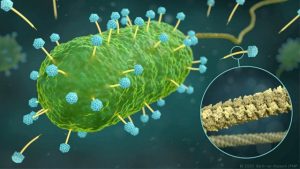 With growing antibiotic resistance, phages have increasingly become the focus of research. Phages are naturally occurring viruses with a very useful property: they implant their DNA into bacteria and proliferate there until the bacterial cell is ultimately destroyed. This is why they are also referred to as bacteriophages (bacteria eaters).
With growing antibiotic resistance, phages have increasingly become the focus of research. Phages are naturally occurring viruses with a very useful property: they implant their DNA into bacteria and proliferate there until the bacterial cell is ultimately destroyed. This is why they are also referred to as bacteriophages (bacteria eaters).
This approach has already been shown to fight multidrug-resistant bacteria. Last year, the case of a girl from England hit the headlines, when she was cured from a serious antibiotic-resistant infection using engineered phages.
However, the widespread use of phage therapy is still a long way off. Many of the underlying principles that are key to advancing this therapy are not yet understood. For example, little was previously known about the appearance of the exact architecture of the tube used by phages to implant their DNA into bacteria. Now scientists from the Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP) in Berlin, together with colleagues from Forschungszentrum Jülich and Jena University Hospital, have managed to reveal the 3D structure of this crucial phage component in atomic resolution.
“The structure and flexibility of the DNA tube attached to the icosahedron-shaped capsid is somewhat reminiscent of a spinal column,” stated FMP’s Professor Adam Lange, describing one of the new findings. “It seems to be perfectly designed for transporting DNA.”
The researchers were able to gain insights into the structure and function of this sophisticated DNA transport pathway – in this case, from a variant of phage SPP1 – by combining solid-state NMR with cryo-electron microscopy (cryo-EM). Lange’s research group further developed nuclear magnetic resonance spectroscopy (NMR) especially for this task under an ERC Grant; cryo-EM expert Professor Gunnar Schröder from Forschungszentrum Jülich performed the electron-microscopic investigations. In addition, new modeling algorithms were required for the computer-based combination of the two data sets for structure determination. These algorithms were developed by Professor Michael Habeck from Jena University Hospital. “The key to success was combining the two methods, representing a methodological milestone,” commented Professor Lange.
While solid-state NMR is ideal for visualizing flexible structures and tiny details, cryo-EM provides insight into the overall architecture. The resulting image shows that six gp17.1 proteins organize into stacked rings, forming a hollow tube. The rings are connected by flexible linkers, making the tube very bendable. “We are now able to understand how negatively charged DNA is repelled from the likewise negatively charged interior wall of the flexible tube, passing through it smoothly,” explained FMP’s Maximilian Zinke, lead author of the study now published in Nature Communications. “The bacteria are ultimately destroyed via this pathway.”