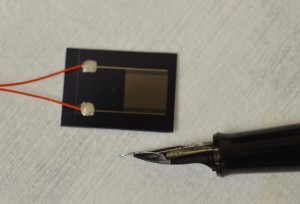
The need for rapid, specific and sensitive assays that provide a detection of bacterial indicators are important for monitoring water quality. Rapid detection using biosensor is a novel approach for microbiological testing applications. Besides, validation of rapid methods is an obstacle in adoption of such new bio-sensing technologies.
 In this study, the strategy developed is based on using the compound 4-methylumbelliferyl glucuronide (MUG), which is hydrolyzed rapidly by the action of E. coli β-D-glucuronidase (GUD) enzyme to yield a fluorogenic product that can be quantified and directly related to the number of E. coli cells present in water samples. The detection time required for the biosensor response ranged from 30 to 120 minutes, depending on the number of bacteria. The specificity of the MUG based biosensor platform assay for the detection of E. coli was examined by pure cultures of non-target bacterial genera and also non-target substrates. GUD activity was found to be specific for E. coli and no such enzymatic activity was detected in other species. Moreover, the sensitivity of rapid enzymatic assays was investigated and repeatedly determined to be less than 10 E. coli cells per reaction vial concentrated from 100 mL of water samples.
In this study, the strategy developed is based on using the compound 4-methylumbelliferyl glucuronide (MUG), which is hydrolyzed rapidly by the action of E. coli β-D-glucuronidase (GUD) enzyme to yield a fluorogenic product that can be quantified and directly related to the number of E. coli cells present in water samples. The detection time required for the biosensor response ranged from 30 to 120 minutes, depending on the number of bacteria. The specificity of the MUG based biosensor platform assay for the detection of E. coli was examined by pure cultures of non-target bacterial genera and also non-target substrates. GUD activity was found to be specific for E. coli and no such enzymatic activity was detected in other species. Moreover, the sensitivity of rapid enzymatic assays was investigated and repeatedly determined to be less than 10 E. coli cells per reaction vial concentrated from 100 mL of water samples.
The applicability of the method was tested by performing fluorescence assays under pure and mixed bacterial flora in environmental samples. In addition, the procedural QA/QC for routine monitoring of drinking water samples have been validated by comparing the performance of the biosensor platform for the detection of E. coli and culture-based standard techniques such as Membrane Filtration (MF). The results of this study indicated that the fluorescence signals generated in samples using specific substrate molecules can be utilized to develop a bio-sensing platform for the detection of E. coli in drinking water. The procedural QA/QC of the biosensor will provide both industry and regulatory authorities a useful tool for near real-time monitoring of E. coli in drinking water samples. Furthermore, this system can be applied independently or in conjunction with other methods as a part of an array of biochemical assays in order to reliably detect E. coli in water.
Biosensor platform for rapid detection of E. coli in drinking water
Arizona State University Digital Repository
Hesari, Nikou / Abbaszadegan, Morteza / Alum, Absar / Fox, Peter / Stout, Valerie