Princeton researchers have for the first time revealed the mechanics of how bacteria build up slimy masses, called biofilms, cell by cell. When encased in biofilms in the human body, bacteria are a thousand times less susceptible to antibiotics, making certain infections, such as pneumonia, difficult to treat and potentially lethal.
 In a study published Sept. 6 in the Proceedings of the National Academy of Sciences, a team at Princeton tracked a single bacterial cell as it grew into a mature biofilm of 10,000 cells with an ordered architecture. The findings should help scientists learn more about bacterial behavior and open up new ways of attacking biofilms with drugs.
In a study published Sept. 6 in the Proceedings of the National Academy of Sciences, a team at Princeton tracked a single bacterial cell as it grew into a mature biofilm of 10,000 cells with an ordered architecture. The findings should help scientists learn more about bacterial behavior and open up new ways of attacking biofilms with drugs.
“No one’s ever peered inside a living biofilm and watched it develop cell by cell,” said Bonnie Bassler, a senior author of the paper and the Squibb Professor in Molecular Biology at Princeton, as well as a Howard Hughes Medical Institute Investigator. “With this paper, we can now understand for the first time how communities of bacteria form a biofilm.”
The discovery became possible thanks to a special microscopy method pioneered at Princeton by a former postdoctoral research associate, Knut Drescher, which allowed the imaging of single cells, letting researchers follow a budding biofilm in real time.
“We have used a state-of-the-art technique to see into the core of a living, growing biofilm,” said postdoctoral research associate Jing Yan, lead author of the new study. Along with membership in Bassler’s lab, Yan belongs to the Complex Fluids Group led by paper senior co-author Howard Stone, the Donald R. Dixon ’69 and Elizabeth W. Dixon Professor of Mechanical and Aerospace Engineering at Princeton. Yan is further advised by paper senior author Ned Wingreen, the Howard A. Prior Professor of the Life Sciences and acting director of the Lewis-Sigler Institute for Integrative Genomics at Princeton.
“The research that produced this paper sits at the frontier between materials science, engineering, physics and biology and represents a fantastic collaboration across Princeton University,” said Bassler.
Along with Yan, Bassler, Stone and Wingreen, a fifth co-author of the paper is Andrew Sharo, a former undergraduate in Princeton’s Department of Physics and now at the University of California-Berkeley.
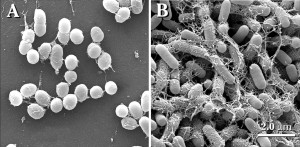
The researchers chose Vibrio cholerae for their model biofilm organism because of its long history of study and threat to human health, causing the diarrheal disease cholera. A curved, rod-shaped bacterium, V. cholerae lives as a free-swimming cell in brackish water or saltwater. When V. cholerae makes contact with a food particle, perhaps on the shell of a crab or a shrimp, or a human intestinal cell during disease, the bacterium attaches itself and begins to reproduce. The expanding colony’s members secrete a glue-like substance to keep from getting washed away and to protect themselves from competing bacteria.
Previous efforts to delve into how the cells in a burgeoning biofilm interact had failed because of insufficient optical resolution; basically, what one cell was doing in the opaque mass could not be distinguished from its neighbors.
The Princeton researchers overcame this problem in several ways. First, they genetically modified the bacterial strain so the cells produced proteins that glow brightly when illuminated by specific colors of light. The proteins selected offer the brightest available fluorescence, making each cell easier to pick out, while reducing the intensity of potentially cell-damaging light required for the experiment.
The team then used a confocal microscope, a device that focuses on a single portion of a specimen from a certain distance. By making hundreds of such observations, images can be stacked together to create a three-dimensional image of the entire specimen. “It’s like looking deep into the interior of a biofilm without having to slice it open,” said Yan.
Another boost for the research team came from computer algorithms originally developed for fields like materials science. The algorithms differentiated closely clustered sources of light, in this case the many bunched-up V. cholerae cells in a thickening biofilm.
What the Princeton team saw was remarkable. At first, the bacterial colony expanded horizontally on the given surface in the experiment. As each cell split, the resulting daughter cells firmly attached to the surface alongside their parent cells. Squeezed by increasing numbers of offspring bacteria, however, the cells at the heart of expanding colony were forced to detach from the surface and point vertically. The bacterial colony thus went from a flat, two-dimensional mass to an expanding, three-dimensional blob, all held together by gunk in the developing biofilm.
The Princeton team dug a bit deeper into the genetics behind this cellular behavior. A single gene, dubbed RbmA, is key to behavior in which new cells connect in such a way to develop a three-dimensional biofilm. When the researchers deactivated the gene, a big, diffuse and floppy biofilm formed. When RbmA performed as normal, though, a denser, stronger biofilm resulted as the cells stayed linked to each other. Thus, RbmA provides the biofilm its resilience, providing insight into a potential Achilles heel that could be targeted for therapeutic intervention.
Ongoing work is now measuring the physical forces experienced by cells uplifting at the biofilm’s center so the overall mechanics can be precisely worked out. “We are currently trying to develop a mathematical model for how the bacterial colony grows in time and how the spatial features are linked to typical mechanical features of the biofilm,” said Stone.
 Three bacterial species selected were assessed using a culture‐dependent approach followed by verification of isolates using both BIOLOG GEN III and polymerase chain reaction. The estimated number of each bacterium was 122 isolates, while 60, 20, 26, and 16 isolates were obtained from the natural biofilm samples, kitchen, bathroom, laboratories, and hospital, respectively. As for the tests, in all types of biofilm samples, the overall bacterial counts at low temperature (22°C) were higher than those at high temperature (37°C). Meanwhile, E . coli had the most significant number of bacterial microorganisms compared to the other two pathogens. Additionally, the most massive cell densities of E . coli , Salmonella , and Listeria species were discovered in the biofilm collected from the kitchen, then the hospital.
Three bacterial species selected were assessed using a culture‐dependent approach followed by verification of isolates using both BIOLOG GEN III and polymerase chain reaction. The estimated number of each bacterium was 122 isolates, while 60, 20, 26, and 16 isolates were obtained from the natural biofilm samples, kitchen, bathroom, laboratories, and hospital, respectively. As for the tests, in all types of biofilm samples, the overall bacterial counts at low temperature (22°C) were higher than those at high temperature (37°C). Meanwhile, E . coli had the most significant number of bacterial microorganisms compared to the other two pathogens. Additionally, the most massive cell densities of E . coli , Salmonella , and Listeria species were discovered in the biofilm collected from the kitchen, then the hospital.